Introduction¶
The Field Programmable Gate-Arrays(FPGAs) cluster offers a powerful hardware platform capable of performing realistic simulations of Biological Neural Systems(BNS). However, making use of such powerful platform is not easy and not all users have enough understanding and experetise to exploit such hardware. Having a user friendly software platform to interact with the FPGAs in a much higher abstraction level will allow more researchers to make use of such powerful hardware without having to spend a lot of time actually understanding and programming the platform. Such user interface takes away a certain level of abstraction and makes it intuitive and easy to generate desired biologically inspired neural network with desired neuron models, synpase models, network size, network topology and desired stimulation configuration for the neural network. Here, we discuss web based user interface that we developed to interface with the FPGA cluster that allows users to easily and intuitively create desired network to be run on the FPGA cluster.
Note
So far only biologically inspired spiking neuron models are implemented.
Overview of the platform¶
The simulator platform is mainly composed of three distinct functional layers - software layer (Web Interface), intermediate layer (Interface Manager(IM)) and hardware layer (FPGA cluster). Software layer is the user interface which acts as front end part of the platform that allows users to interact with the platform.
The web interface is linked to the interface manager (IM) which works as an intermediate node between web interface and the FPGA clusters and manages flow of packets to and from both FPGAs and web interface. IM receives the simulation initialisation files, which contains all the information about the simulation in the xml format, from the web interface generated by the users which are then sent to the appropriate FPGA nodes for simulation. Once the simulation has completed, the results packets are sent by the FPGA clusters to the IM with correct information of simulation and user for correct redirection. IM then handles the incoming packets from the FPGA cluster and send it to the users.
The simplified version of the workflow can be shown as below:
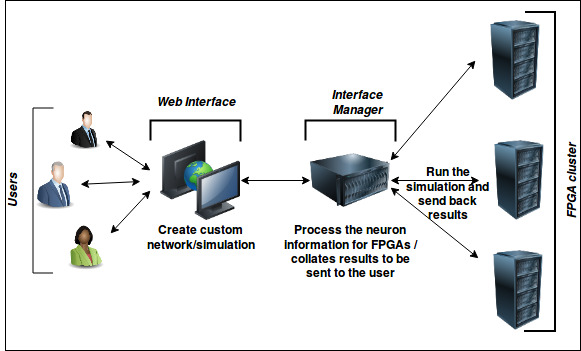
Figure1: Simplified work flow of the whole process
Some of the key features of the web interface are listed below:
- Custom network of spiking neurons with parameterisable simulation and models with custom stimulation parameters.
- Parameterisable neuron models from the model library(Current Models: Integrate and Fire, Leaky Integrate and Fire, Izhekevich).
- User specification of neurons with same or different neuron models. If you want same models with different paramters then you can also do that.
- Selection of layered or non-layered network. Layered network would be a feedforward network where the first layer is the input layer and last layer is the output.
- Selection of same or different neruon models on each layer. All the neurons in a single layer will have the same neuron model.
- Topology selection; fully interconnected, random or probabilistic(30% connection at the moment)
- Simulation parameter configuration, such as simualtion time, timestep size, watchdog period etc.
- Stimulation parameter configuration such as start and end time and voltage. [Only step stimulation is implemented at the moment]
- Visualisation of the spike train obtained after the simulation as raster plot.
Requirements¶
The following requirements should be met:
- Mozilla or Google chrome browser (Recommended).
- CPU requirements
Disclaimer¶
- The contents of the platform remains a sole Intellectual property of Nottingham Trent University.